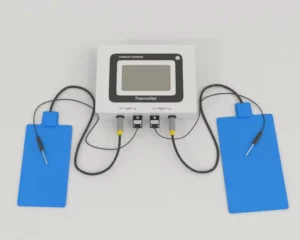
Products, services, boutique tech transfer
TOOLS TO
CONDUCTSCIENCE
GET PUBLISHED, FOR LESS
Trusted by INNOVATIVE Organizations





Our OFFERINGS
Free Shipping
For Local Orders Over $1,000**
Exchange Offers
Free returns within 30 days
Bulk Purchases
Bulk orders, more for less
Help & Support
Contact Our Team 24/7
Our SCIENTIFIC Services
Explore our services categories clicking below
RESEARCH LAB
VIRTUAL REALITY
Unlock Virtual Reality for Scientific Experiments
Simian Virtual Reality is a web-based publisher for scientific experiments. Your experiment is built for you, the data is generated and outputted to you in easy-to-publish formats.
Simple, Easy and Flexible
Starting from $4,900

SPECIMEN LAB
MICROSCOPES
Microscopy plays a crucial role in scientific research. Its high-resolution imaging capabilities allow scientists to observe and analyze samples at the microscopic level. By studying the structure and behavior of cells, tissues, and materials, researchers gain valuable insights into disease mechanisms, material properties, and biological processes.
Starting from $1,200
ANIMAL LAB
SYRINGE PUMPS
Syringe pumps are essential tools in scientific research and experimentation, providing precise and controlled delivery of fluids with exceptional accuracy and repeatability. Whether you are involved in pharmacology, neuroscience, biotechnology, or any other field that requires accurate fluid delivery, our syringe pumps are designed to meet your needs.
Starting from $1,390

FEATURED Categories
Explore our best product categories clicking below
Our best PRODUCTS
By categories / best sellers
- Stereotaxic
- Stereotaxic Accesories
- Stereotaxic Ear Bars
- Stereotaxic Holders
- Restrainers
- Animal Cages & Restrainers
- Surgical Equipment
- Lab Consumables
-
$185.00 – $195.00
-
$1,850.00 – $1,990.00
-
$280.00 – $405.00
-
$16,900.00 – $17,100.00
-
$270.00
Best SELLERS
-

Rodent Heating Pad
$185.00 – $195.00 Select options -

LCD Screen Laboratory Constant Syringe Pump
$1,390.00 – $1,590.00 Select options -

Standard Stereotaxic Manual Surgery Instrument for Rat
$3,100.00 Add to cart -

Stainless Steel Rabbit Restrainer
$530.00 Add to cart -

Diffusion Cell Apparatus
$2,800.00 Add to cart -

Standard Manual Stereotaxic for Rat and Mouse
$4,390.00 Add to cart -

CO2 Incubator
$3,900.00 – $4,900.00 Select options -

Inverted Fluorescent Microscope
$6,690.00 Add to cart